Research
Success is not final, failure is not fatal. It is the courage to continue that counts. By Winston Churchill.
Our research focuses on developing and applying novel statistical and computational methodologies to tackle complex challenges in genetics, genomics, and biomedical science. We leverage LLM, DNA foundation models, causal inference, machine learning, and integrative multi-omics analysis to gain deeper insights into drug target discovery, clinical trial improvement, disease mechanisms, identify biomarkers, improve risk prediction, and enhance drug discovery.

AI for Science
We leverage machine learning and artificial intelligence to advance scientific discovery. Our research includes developing and benchmarking deep learning models for genomics (e.g., DNA foundation models; Feng et al., Nature Communications 2025), applying large language models to clinical data, exploring AI for drug discovery, and creating efficient algorithms for tasks like penalized regression-based clustering (PRclust; Wu et al., JMLR 2016). These approaches aim to extract deeper insights and predictive power from complex biological and clinical data.

Causal Inference and Mendelian Randomization
We develop novel methods for causal inference from observational data. A major focus is Mendelian Randomization (MR), using genetic variants as instrumental variables to probe causal effects while minimizing confounding. We have developed novel techniques to address key MR challenges like the winner's curse bias arising from IV selection (RIVW; Ma et al., Annals of Statistics 2023) and widespread pleiotropy (CARE; Xie et al., arXiv 2023). Beyond MR, our work includes efficient estimation of Heterogeneous Treatment Effects using iterative Targeted Maximum Likelihood Estimation (iTMLE; Wei et al., Biometrics 2023) and robust, nonparametric Propensity Score Estimation through deep learning that directly optimize covariate balance (Peng et al., arXiv 2024).
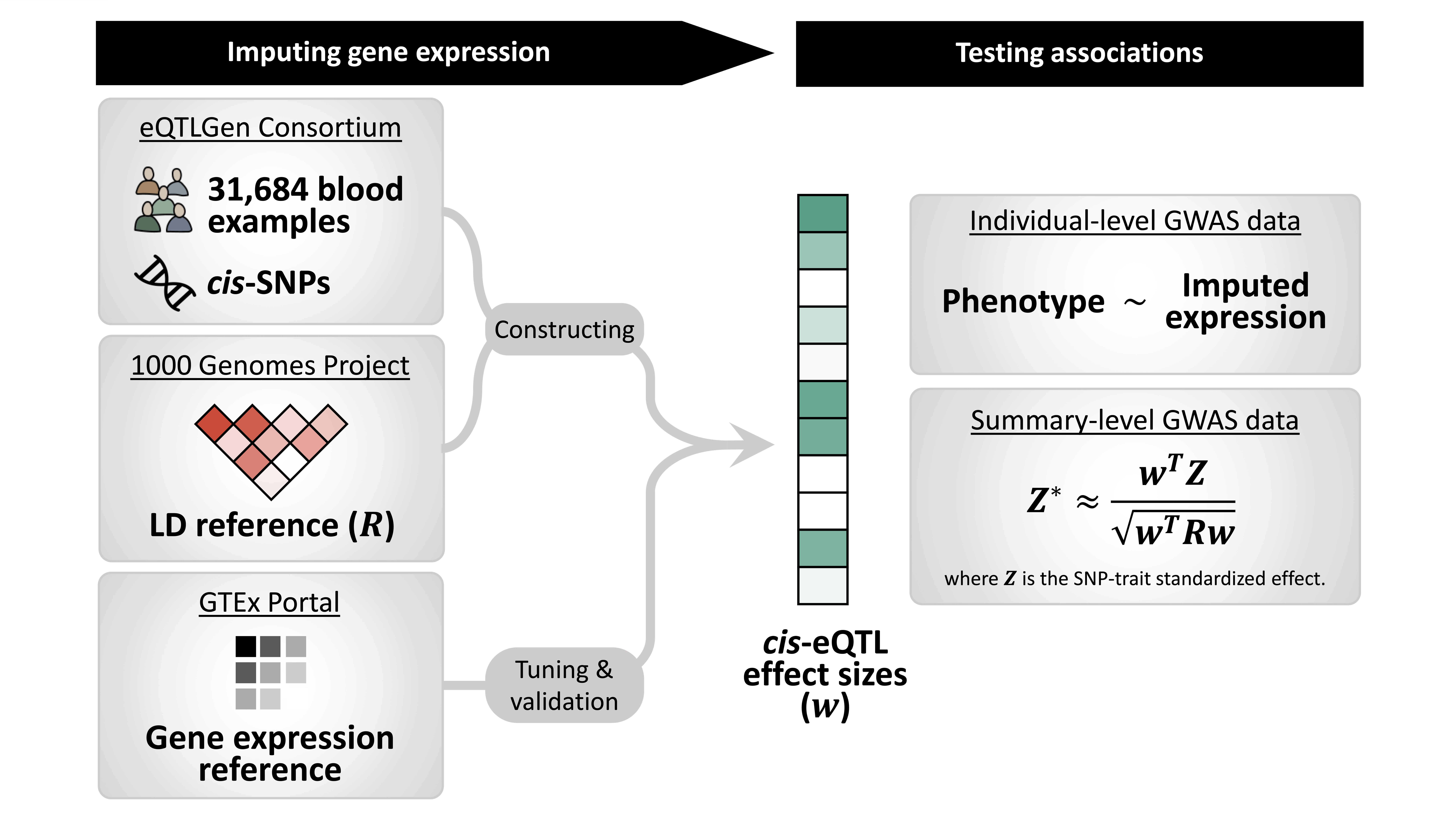
Statistical Genetics and Integrative Analysis
Our group pioneers statistical methods for integrating diverse high-dimensional data (GWAS, eQTL, epigenomics, proteomics) to elucidate disease mechanisms. We develop computationally efficient approaches for multi-omics integration, including advanced imputation techniques (SUMMIT; Zhang et al., Nature Communications 2022), incorporating regulatory elements like enhancers and mQTLs (Wu & Pan, Genetics 2018; Bioinformatics 2019), and powerful fine-mapping strategies (FOGS; Wu & Pan, Human Genetics 2020). These methods aim to identify novel disease-associated genes and pathways from genetics and genomics data.
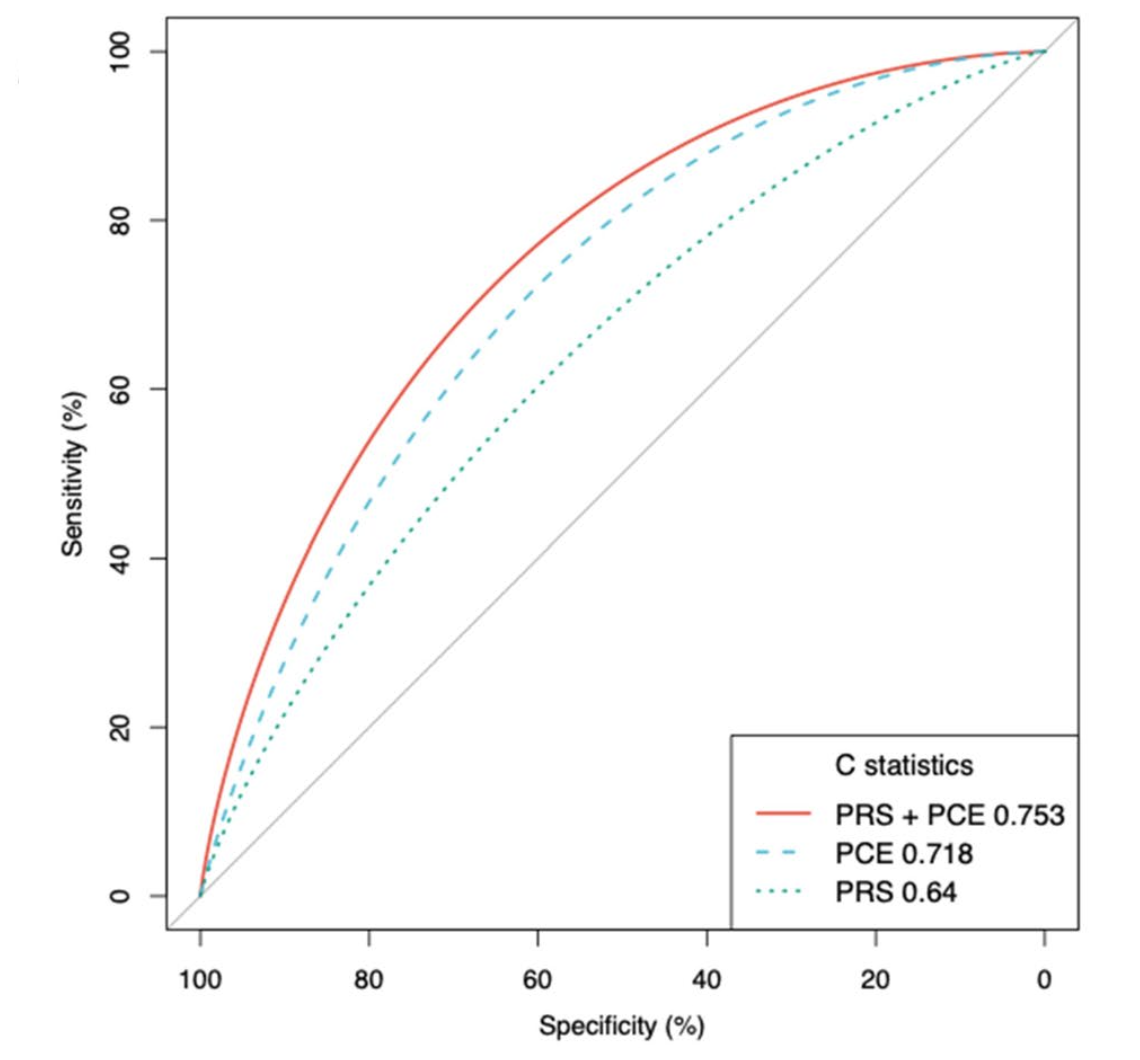
Disease Risk Prediction
A key focus is enhancing disease risk prediction using Polygenic Risk Scores (PRS). We develop methods to integrate multi-omics data and genetically predicted biomarkers with PRS (Wu et al., Cancer Communications 2021) and evaluate strategies to improve the accuracy and clinical utility of PRS for complex diseases like coronary artery disease, comparing performance against standard clinical risk models (King et al., BMC Medicine 2022). Our goal is to contribute to more personalized prevention and early detection strategies.

Applications and Collaborations
Our methodological work is deeply integrated with real-world biomedical research through close collaborations with epidemiologists, geneticists, and clinicians at MD Anderson and other institutions. These partnerships have led to impactful studies identifying putative causal genes for COVID-19 severity (Wu et al., Genetics in Medicine 2021), improving risk prediction for prostate cancer (Wu et al., Cancer Communications 2021), and advancing our understanding of Alzheimer's disease (Wu et al., Bioinformatics 2021a) and pancreatic cancer (Liu et al., Cancer Research 2020). This ensures our statistical tools address critical scientific needs.

Software and Resource Development
A cornerstone of our research is the commitment to open science and reproducibility. We actively develop and maintain robust, validated, open-source software implementing our novel methods, primarily as R packages (e.g., MiSPU, GLMaSPU, prclust, FOGS, CMO). We prioritize user-friendliness, providing comprehensive documentation and tutorials to make complex statistical tools accessible. An example is the Global Causal Biomarker Hub (GCB Hub), a comprehensive resource offering pre-computed biomarker prediction models (proteins) across diverse ancestries and protein-trait associations across several biobanks such as MVP and Finngene. Our group's software and detailed resources are available on our Software page.
High-Dimensional Statistical Inference
We develop theoretically sound and powerful methods for statistical inference in high-dimensional settings (p >> n). Key contributions include adaptive tests for high-dimensional parameters in GLMs, accommodating both low- and high-dimensional nuisance parameters (aiSPU; Wu et al., Statistica Sinica 2019; JMLR 2020), and novel testing procedures based on asymptotically independent U-statistics (He et al., Annals of Statistics 2021). These methods enhance statistical power and control Type 1 error rates in complex genomic and biomedical analyses, including applications to microbiome data (Wu et al., Genome Medicine 2016).
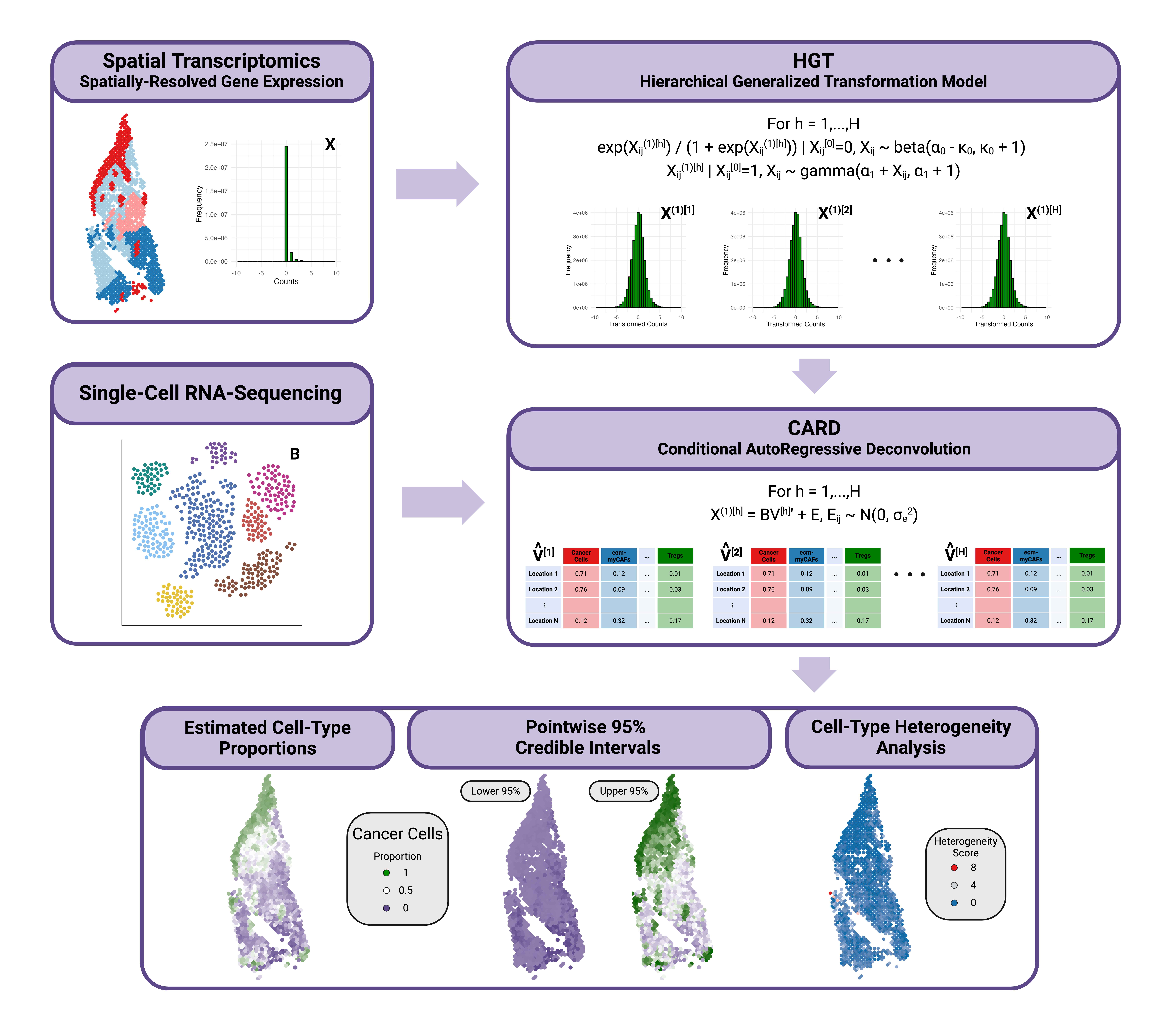
Other Research Areas
Beyond the core areas above, our research extends to developing advanced statistical methodologies for meta-analysis, enabling robust evidence synthesis across studies (e.g., Meng et al., Statistics in Medicine 2024). We are also actively engaged in creating novel analytical approaches for spatial transcriptomics data, focusing on delineating complex tissue heterogeneity, understanding cell-cell interactions, and improving spatially-informed cell-type deconvolution (e.g., Lyu et al., Bioinformatics 2024; Melton, Bradley & Wu, bioRxiv 2024).